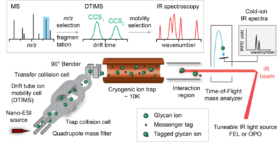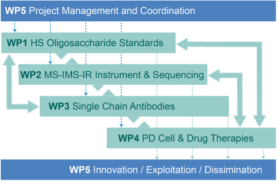
Heparan sulfate (HS) are highly sulfated polysaccharides that reside on the cell surface and extracellular matrix of all mammalian cell types. Understanding their functions has tremendous potential to unlock diverse biomedical applications. Harnessing this promise has been difficult due to a fundamental technology bottleneck – lack of methods for the sequencing of HS structures to define the natural “HS codes” that underpin many biological functions. Availability of effective, rapid methods will open the floodgates to identification and characterization of functional domains leading to the next-generation of HS-based therapeutics and new diagnostics. It is our vision that robust sequencing methods for heparin and HS will only become possible when large collections of fully-defined HS-oligosaccharides standards are made available. HS-SEQ will address this need by preparing such collections of compounds by chemoenzymatic synthesis and fractionation (WP 1). The resulting HS standards will be crucial to developing a structural identification method based on multidimensional metrics such as collisional cross sections (CCS) measured by ion mobility mass spectrometry (IM-MS) and optical response determined by gas-phase infra-red (IR) ion spectroscopy (WP 2). An integrated MS-IMS-IR approach will revolutionize the sequencing of HS-oligosaccharides obtained from complex biological mixtures, permitting a step-change in structure-activity studies. The HS-oligosaccharide standards will also be exploited to develop an antibody toolkit to identify epitopes in the context of cells and tissues (WP 3). Specific transformational applications of the technology will be pursued: (i) identification of HS codes that promote the generation of dopaminergic neurons from human pluripotent stem cells (hPSCs) for cell replacement therapy in PD; and (ii) to achieve unprecedented in-depth analysis of pharmaceutical heparin to advance quality control and to facilitate identification of novel therapeutic species of next-generation heparins (WP 3).
WP1: Chemical standards and advanced separation methods
HS-SEQ will exploit chemoenzymatic approaches to prepare panels of HS oligosaccharides. To speed up the synthesis, automation protocols will be implemented in which enzymatic reactions are performed in solution, and compound purification is performed by solid phase extraction. In parallel, Enhanced separation technologies will be developed to create a step-change in advanced analysis of heparin/HS oligosaccharides, supported by the synthetic standards, and exploiting novel multiple orthogonal LC workflows . In particular, strong anion exchange will be enhanced significantly through optimised bead coating and quaternary amine spacing.
Responsible/supervisor: UU & UCPH
WP2: MS-IMS-IR spectroscopy for HS sequencing
HS-oligosaccharides can occur in many isomeric forms (same molecular weight but different sulfation and epimerizing patterns) and therefore MS alone cannot readily identify sequences. To address this shorcoming, we will use existing IM-MS and cold-ion IR spectroscopy instrumentation to produce a database of reference CCS and IR fingerprints of HS standards already available and produced in WP1. We will develop a novel, user-friendly instrument prototype in which IM-MS and cold-ion IR spectroscopy are combined. Technically, this will be achieved using an existing, commercially available Waters Synapt G2S IM-MS as platform. To implement capabilities for cold-ion IR spectroscopy, we will use the messenger tagging spectroscopy approach in which m/z-selected oligosaccharide ions and their fragments are cooled in a cryogenic ion trap. To accurately identify large HS oligosaccharides (dp 6-12), the reference data and the MS-IMS-IR technology will be combined within a coherent workflow. To identify unknown HS-oligosaccharides, samples will be separated via LC and infused into the new instrument, where CCSs are measured in the IM-MS section and IR fingerprints are obtained in the cold-ion trap via tagging spectroscopy. The resulting multidimensional datasets will contain i) m/z information, ii) CCS data and iii) highly diagnostic IR fingerprints of the investigated HS oligosaccharide and all its fragments.
Responsible/Supervisors: FU Berlin, UCPH & Waters
WP3: Single chain antibody took kit: selection, characterisation and application
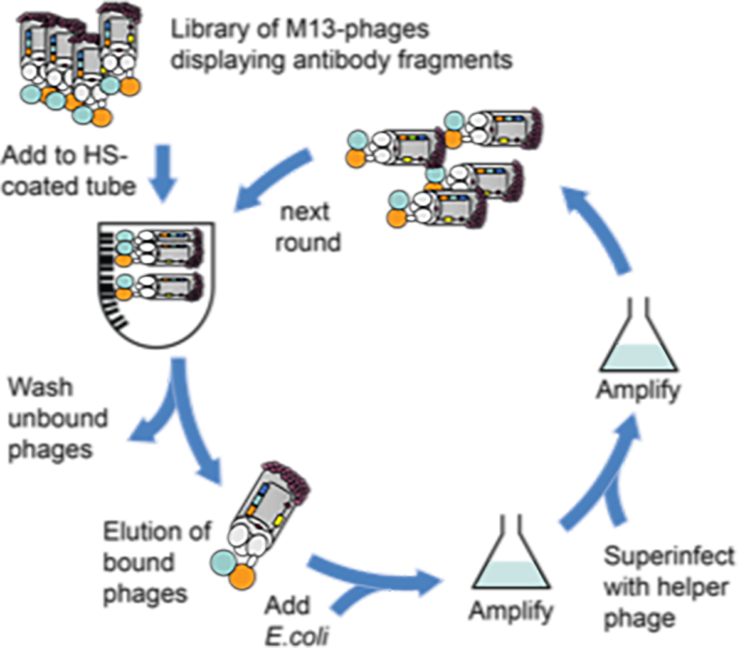
HS-SEQ will develop a large panel of phage-display antibodies for which epitope requirements are fully characterised for the first time. The technological breakthrough here will be twofold: firstly, synthetic HS oligosaccharides will be used to develop new antibodies that selectively recognise epitopes of interest. Secondly, they will be used to fully characterise the epitope requirements of existing antibodies. We envisage a research strategy in which MS-IMR-IR will identify structural differences in the context of healthy/diseased cells, which will guide antibody development to provide a tool to map epitope expression in disease development, and also provide target HS saccharides as future new drugs.
Responsible/Supervisors: RadboudUMC, UU, KI
WP4: Exploiting integrated technologies: PD treatment and pharmaceutical herapins — KI
We will provide proof-of-concept of biomedical application of the HS-sequencing technologies on two different levels. Firstly, the newly developed techniques will used to screen for natural HS saccharides for their ability to promote differentiation of hPSCs to pure mDA neurons, for in-vivo testing as cell replacement therapy for PD. 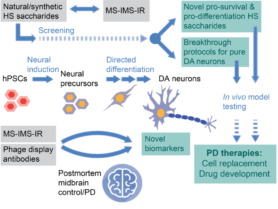 Producing high quality mDA neurons represents a major current technological hurdle. Secondly, the sequencing technologies will also be applied to pharmaceutical and modified heparin to improve QC and harness specific biomedical properties for next-generation herapin drugs.
Producing high quality mDA neurons represents a major current technological hurdle. Secondly, the sequencing technologies will also be applied to pharmaceutical and modified heparin to improve QC and harness specific biomedical properties for next-generation herapin drugs.
Responsible/Supervisor: KI, UCPH, FU Berlin, IntelliHep, UU, RadboudUMC, Opocrin SpA & Waters